Kinetic Modulation of Amyloid-β (1-42) Aggegation and Toxicity by Structure-Based Rational Design
| author | Hugh I. Kim |
|---|---|
| Homepage | https://www.hughkimlab.com/ |
| journal | Journal of the American Chemical Society |
Several point mutations can modulate protein structure and dynamics, leading to different natures. Especially in the case of amyloidogenic proteins closely related to neurodegenerative diseases, structural changes originating from point mutations can affect fibrillation kinetics. Herein, we rationally designed mutant candidates to inhibit the fibrillation process of amyloid-β with its point mutants through multistep in silico analyses. Our results showed that the designed mutants induced kinetic self-assembly suppression and reduced the toxicity of the aggregate. A multidisciplinary biophysical approach with small-angle X-ray scattering, ion mobility-mass spectrometry, mass spectrometry, and additional in silico experiments was performed to reveal the structural basis associated with the inhibition of fibril formation. The structure-based design of the mutants with suppressed self-assembly performed in this study could provide a different perspective for modulating amyloid aggregation based on the structural understanding of the intrinsically disordered proteins.
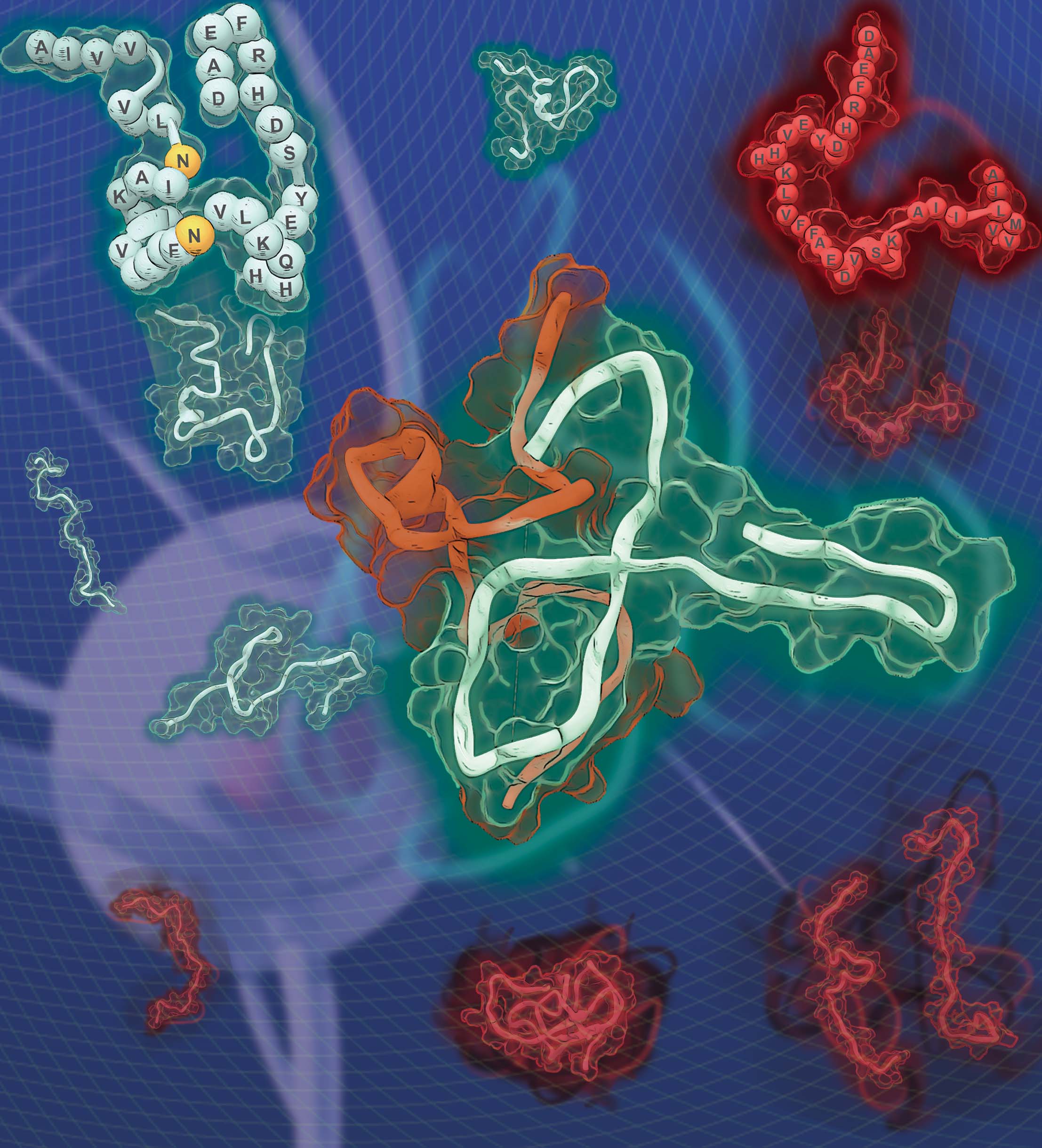
https://pubs.acs.org/doi/10.1021/jacs.1c10173

Lim Dongjun
(First author, integrated course student)
Articles
-
 Highly Efficient Fullerene-Free Polymer Solar Cells Fabricated with Polythiophene...
Highly Efficient Fullerene-Free Polymer Solar Cells Fabricated with Polythiophene...
-
 Low-voltage organic devices based on pristine and self-assembled monolayer-treate...
Low-voltage organic devices based on pristine and self-assembled monolayer-treate...
-
 Higher Quantum State Transitions in Colloidal Quantum Dot with Heavy Electron Doping
Higher Quantum State Transitions in Colloidal Quantum Dot with Heavy Electron Doping
-
 High-harmonic generation: Drive round the twist
High-harmonic generation: Drive round the twist
-
 Gradients of Rectification: Tuning Molecular Electronic Devices by the Controlled...
Gradients of Rectification: Tuning Molecular Electronic Devices by the Controlled...
-
 Investigation of Charge Carrier Behavior in High Performance Ternary Blend Polyme...
Investigation of Charge Carrier Behavior in High Performance Ternary Blend Polyme...
-
 Adsorption of Carbon Dioxide on Unsaturated Metal Sites in M2(dobpdc) Frameworks ...
Adsorption of Carbon Dioxide on Unsaturated Metal Sites in M2(dobpdc) Frameworks ...
-
 Programmed activation of cancer cell apoptosis: A tumor-targeted phototherapeutic...
Programmed activation of cancer cell apoptosis: A tumor-targeted phototherapeutic...
-
 Probing Distinct Fullerene Formation Processes from Carbon Precursors of Differen...
Probing Distinct Fullerene Formation Processes from Carbon Precursors of Differen...
-
 Manifesting Subtle Differences of Neutral Hydrophilic Guest Isomers in a Molecula...
Manifesting Subtle Differences of Neutral Hydrophilic Guest Isomers in a Molecula...
Designed by sketchbooks.co.kr / sketchbook5 board skin